Protein-Ligand Binding Studies
In early discovery or lead optimisation phases, the binding of the target by the candidates is the key parameter for functional analysis. Weak binders are often most attractive – they have the basic interaction at the binding site. However, there is plenty of scope for optimising the structure to increase the potency and specificity of the target. As a molecule passes through rounds of optimisation, reassessment of the binding can inform further work. Many available technologies can assess the binding of a small molecule to a protein. In most development programs, several are used to give an orthogonal view and, therefore, greater confidence. Each technology has strengths and weaknesses, and choice tends to be a personal preference by a researcher with various approaches to offer the richest dataset.
Challenges of Ligand Binding
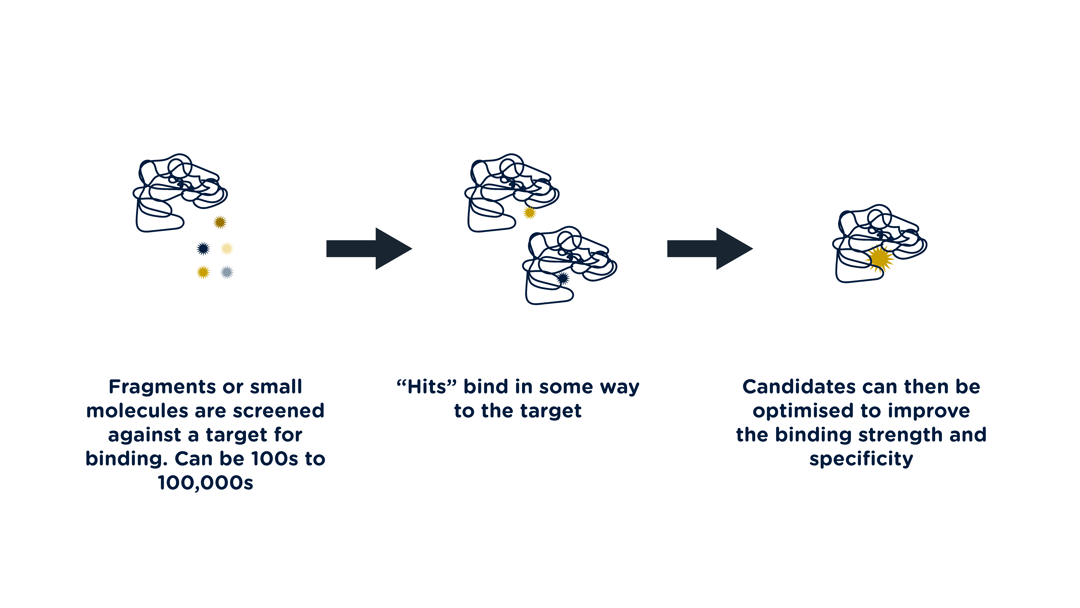
Volume is one of the main challenges of ligand binding, at least in the early screening phase, where hundreds of thousands of molecules may be screened. Typically, a fragment screen is much lower than this; however, it brings a further challenge of weaker binding, which some techniques can miss. Rapid turnaround is also an advantage for the optimisation steps. One of the techniques that can be employed is thermal shift analysis, the binding of the ligand to the protein causing a stabilising effect which can be detected by measuring the Tm shift. Over a range of concentrations, this shift can be used to calculate the dissociation constant to reveal the strength of binding.
Interactions analysis by the SUPR-DSF offers measurements across a wide analyte binding concentration range; it is perfectly suited to see weaker binding interactions than most other techniques. The measurements are free in solution and are not influenced by surface effects, mass transport or refractive index buffer issues, thus providing the perfect complementary data to more traditional binding analysis techniques. Connectivity with other techniques is important here as samples can be prepared in the same formats, such as 384-well plates, and high throughputs are achievable to ensure this part of the screen is not the rate-limiting step.
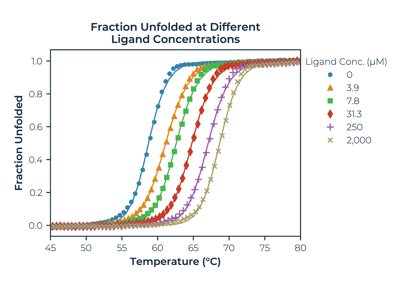
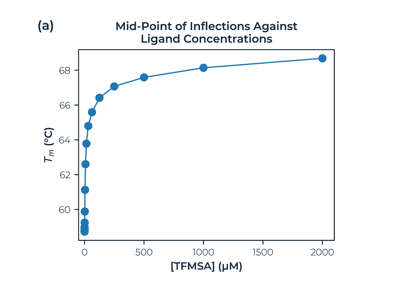
Fraction unfolded melting curves of Human Carbonic Anhydrase with ligand TFMSA at increasing concentration. The second plot shows the effect of concentration on the melting temperature.






