Protein Engineering and Construct Screening
Once a target of interest has been identified, the first stage of a large molecule discovery program is to find or create molecules of interest that have some level of effect on that target. Multiple ways exist, from the more “shotgun” phage display to a more targeted point mutation strategy, resulting in construct libraries of various sizes. Function is key – for example, does the molecule bind to the target? It does not matter how weak that binding may be, as that can be addressed downstream in an optimisation program. Another important factor is the molecule's stability, which indicates how likely the construct will move through the discovery pipeline and perhaps eventually become a marketed molecule. Having the most stable form of the protein, for example, an engineered antibody is a solid basis for further optimisation work before forwarding the candidates to the next development steps.
Challenges of Construct Screening
Often involving hundreds to thousands of early constructs, all of which have been produced in very small quantities, screening for stability requires higher throughputs, low sample usage and the ability to link seamlessly to other parts of the process, such as liquid handling and automation.
Using the SUPR-DSF from Protein Stable is the natural choice for construct stability screening with direct in plate measurements. This means that 384-well microplates can be used for sample preparation, and the plates can be directly processed in the SUPR-DSF. The rank order of stabilities can be produced in a matter of hours, not days and the data-rich spectra are used to predict anomalies such as self-association in addition to producing Tm.
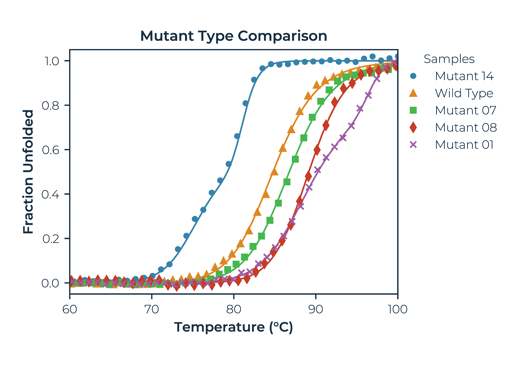
Simple protein thermal melt analysis, ranking mutants against a wild-type. In this example, Mutant 14 destabilises and is not a good candidate. Mutant 01 increases stability the most and introduces a second transition to the melting profile. Mutant 08 is the strongest candidate with the largest increase in single transition Tm.






