Speeding Up Early Stage Biotherapeutic Discovery
Introduction
Limited amounts of sample often hinder the analyses needed for the selection and optimization of the candidates in the early stages of biotherapeutic discovery. Examples of analyses are activity (often binding the target and causing a change in the response to normal function) and stability (potential to survive both the manufacturing process and longer-term storage).
Existing technologies for measuring stability of biologics have all proved useful in identifying candidates that have higher stability than others under certain parameters but have been compromised in some way. There are technologies which give excellent results but may be slow and unsuitable for larger numbers of samples. Others can work on higher numbers of samples, but perhaps lose some data integrity or use too much sample to be a viable option. The SUPR-DSF from Protein Stable eliminates these challenges and combines low sample usage and highly informative data on the stabilities of thousands of samples. The use of the industry-standard format of 384-well microplates eliminates the expensive and risky steps of manual handling by combining sample preparation and analysis in the same plate. Fitting seamlessly into existing processes, stabilities of thousands of samples can be measured and compared in a matter of hours, not days and weeks.
To illustrate the use of the SUPR-DSF for variant comparison and selection in early-stage discovery, we have used a model protein system to compare 16 analogues[1] and identify the most stable ones for further processing.
Results
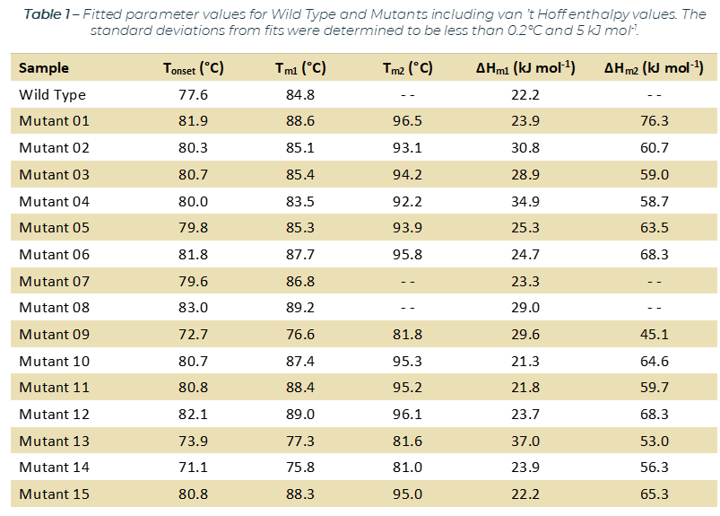
The SUPR-DSF produced high quality melting curves for all 16 samples, which included a Wild Type (WT) protein and 15 single-point mutation variants. The calculated melting temperatures agreed to within 0.2°C for the triplicate data. The fraction unfolded curves for all 15 mutants (Orange) compared to WT (Blue) are in Figure 1. Three of the mutants (9, 13, and 14) decreased the stability of the WT protein, which is easily seen by the shift to the left in the melting curve. The remaining 12 mutants all increased the stability of WT protein with Mutants 1, 8, and 12 showing the largest increase in stability.
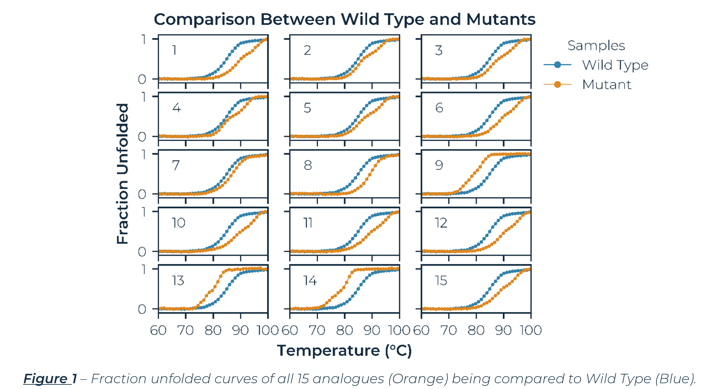
Figure 2 below, illustrates the fraction unfolded melting curves for a representative subset of samples measured. A closer look at the melting curves reveals that some proteins melt via a single transition, while others clearly show two transitions. The WT protein along with Mutants 7 and 8 melt in a single smooth transition, but Mutants 1 and 14 melt via two transitions as shown by two inflection points in the melting curve. Mutants with two transitions include both more stable mutants (Mutant 1) as well as less stable mutants (Mutant 14). In addition, several of the mutants, including Mutant 1 in Figure 2, followed a very similar melting curve to Mutant 8 for the first transition, but then appear to stabilize a second transition at higher temperatures. The ability of mutations to change the protein unfolding from a single to a double transition suggests that additional modifications should be focused to the less stable regions of the protein. The complete results for all the analogues are listed in Table 1. Overall, there are several promising candidates to further investigate to understand why certain mutations are improving the stability of the WT construct.
Conclusion
The SUPR-DSF produced high quality melting curves of 16 analogues or 48 samples (16 x 3 replicates) in 1.5 hours, however this could be easily extended to 384 samples in the same timeframe as the rate-limiting step was running an 80oC ramp at 1oC per minute. This is a significant increase in throughput compared to current methodologies for thermal ramping stability studies. The system recorded not only melting temperature data, but also onset temperatures for unfolding and van ’t Hoff enthalpies, giving deeper insight into the differences between variants. Even small differences could be determined between mutants with limited sample amounts due to the sensitivity of the SUPR-DSF system. Such insights can avoid downstream failures of promising candidates, thus saving time and costs in the biotherapeutic discovery process.
Methodology
The samples were prepared by automatically dispensing the analogues and buffer into a commercially available 384-well microplate at final concentration of 0.2 mg⁄mL and a total well volume of 20 µL. In total, 16 samples in triplicate (48 wells) were dispensed. A commercially available film was placed on the microplate to prevent evaporation, and the plate was ramped from 20°C - 100°C at a ramp rate of 1°C per minute. The data was fit to standard equations[2], and analogues were ranked by examining their melting temperatures. All results and methods are available on request.
References
[ 1] – We thank Romas Kazlauskas’s laboratory (University of Minnesota) for the 16 analogues used in this study.
[2] – J. Walters, S. L. Milam, A. C. Clark. “Practical Approaches to Protein Folding and Assembly: Spectroscopic Strategies in Thermodynamics and Kinetics”. Methods in Enzymology. Vol. 445, p. 1‑39. 2009.