Targeting Optimal Buffers for Downstream Crystallisation Screening
Introduction
Elucidation of protein structure is required to fully understand protein mechanisms of action and function. Structural studies initially require purification of recombinant proteins that are subsequently used for downstream experiments. Protein structure can be investigated using techniques including X-ray crystallography, NMR and cryo-EM. Secondary and tertiary structure can be investigated with circular dichroism and mass spectrometry. Protein purification holds unpredictable challenges, each protein is unique in the conditions required for it to exist in a homogenous form. Buffer composition is one of the strongest influencers of this. Buffers can be optimised to maintain protein homogeneity, increase solubility and stability, and prevent protein unfolding and aggregation, characteristics critical to successfully elucidate structure using the above methods.
The process of X-ray crystallography requires purified protein to form crystals in optimal buffer conditions on specialised crystallisation plates. A primary buffer screen of larger global parameters (buffer system, pH, salt, concentration etc.) before this experimental step can greatly reduce the window of buffers to test and minimise consumable costs. Proteins that easily aggregate or show instability can have their conditions optimised from a preliminary buffer screen before crystallisation trials. The stability of proteins in initial buffer conditions has been directly linked with crystal formation success in subsequent crystallography screens [1]. One method of measuring protein stability in a buffer screen is with differential scanning fluorimetry (DSF). Comparison of thermal melting profiles of a protein in different buffers can indicate the conditions that increase protein stability. We show how the SUPR-DSF can be used to rapidly screen bovine Beta‑lactoglobulin (BLG) in a variety of buffer conditions to increase stability and subsequently aid in improving homogeneity in solution (Figure 1). The system’s 384-well plate format allows simple screening of commercially available buffer screens, plus replicates, in one thermal ramp experiment. These screens have minimal set-up time and require no extrinsic dyes. A preliminary buffer screen can be used to ensure proteins are stored in optimal buffer conditions prior to any structural methodologies being implemented.
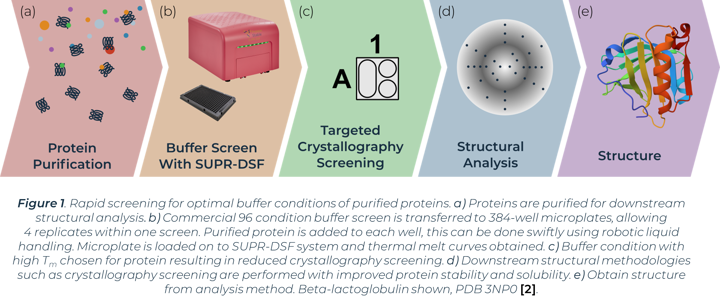
Results
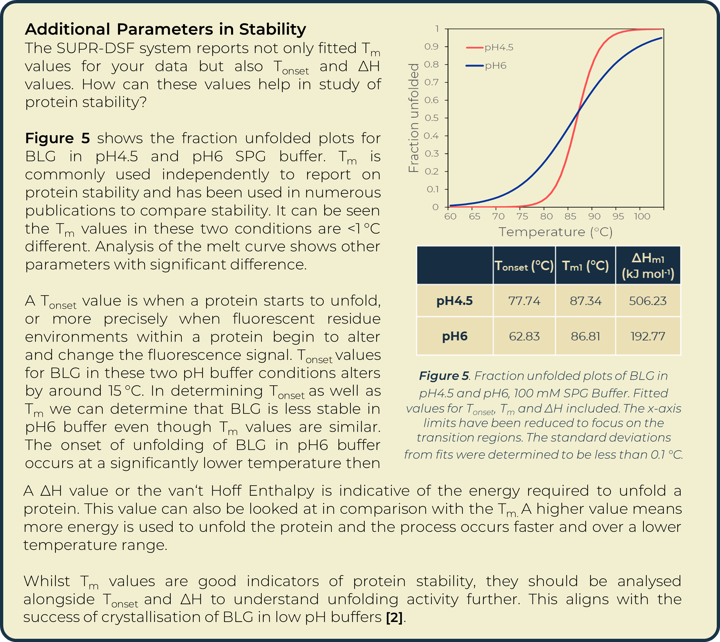
A thermal melt experiment of BLG was performed on the SUPR-DSF using the 96 condition RUBIC buffer screen from Molecular Dimensions [3]. This screen covers large global parameter changes to buffers. Screening of BLG in these conditions showed significant changes in stability dependent on buffer components and type that can be seen within Figure 2. An increase in Tm shows an increase in stability of BLG, these conditions are more suitable for protein storage and downstream protein crystallography screening. A lower Tm shows a decrease in stability and subsequent increased propensity for aggregation.
Heat map analysis of 96 conditions allows for quick comparison of buffer conditions and can easily be related to the suppliers’ buffer screen plan when replicates pipetted in parallel as seen in Figure 2a and 2b. In plotting ΔTm to the control we can see how buffer composition alters stability of proteins negatively or positively. Buffer type (with or without salt) shows higher stability of BLG in conditions that correlate with Citric Acid buffer. The ionic effect of buffer concentration shows little significance in positively altering stability. In most examples these lowered stability of the protein compared to the BLG control. Increased concentrations of imidazole (from well O15 onwards) showed significant destabilizing effects on BLG.
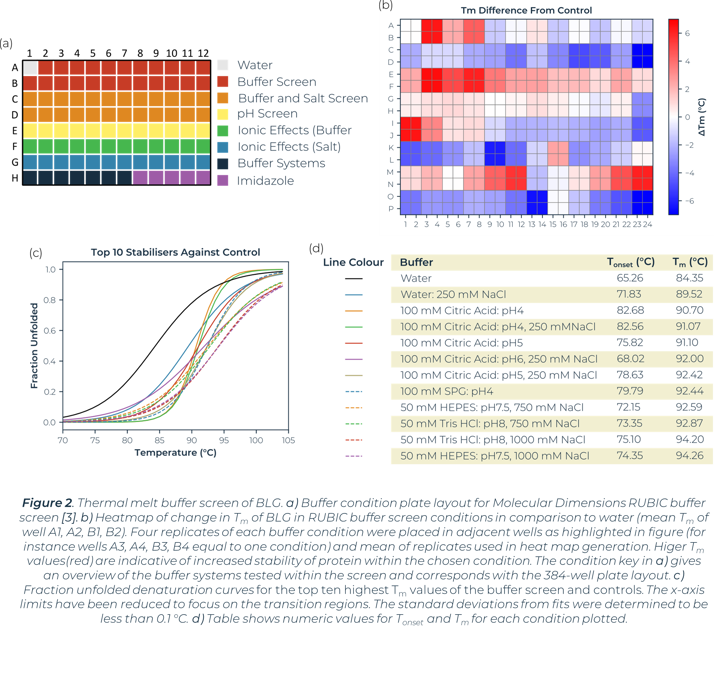
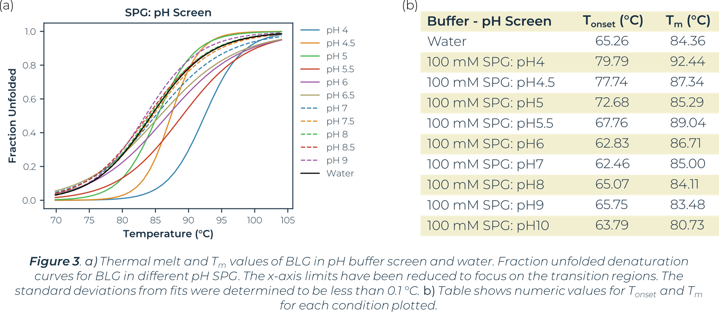
Utilizing the heatmap, we can rapidly identify that salt and pH independently have the largest effect on BLG stability. Identifying the highest Tm values shows that citric acid buffers, which maintain low pHs, are present for half of the top ten stabilizing buffer conditions. SPG (succinic acid, sodium dihydrogen phosphate and glycine in a ratio of 2:7:7) is employed to screen pH effects. The components differing buffering abilities means a large range of pH can be screened without altering the underlying buffer composition. The effects of pH on a protein can then be tested independently of buffer system. The heat map analysis of this pH screen (L1 onwards) shows a correlation of BLG stability and pH change, one of the top ten highest Tm values is in the most acidic condition tested, pH4. Fraction unfolded plots of BLG over a pH range show significant changes in stability. Lower pH values show an increased Tm and Tonset value indicative of a positive correlation between acidity and BLG stability, seen in Figure 3.
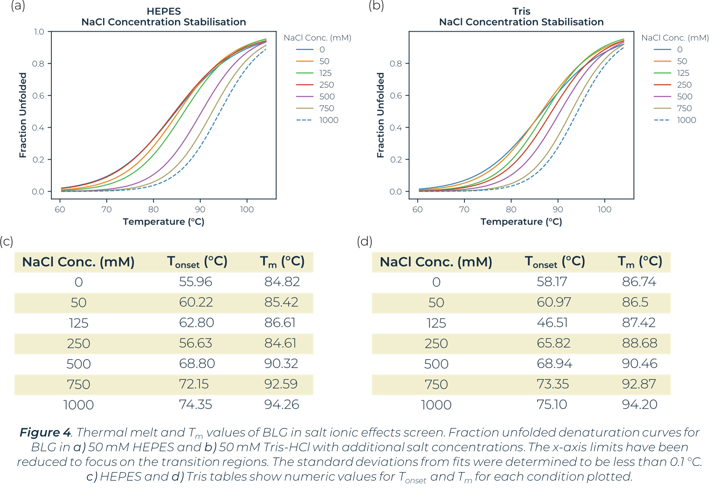
In the global buffer screen, the top stabilising Tm values are within the plate wells measuring the ionic effects of salt. In this instance both Tris-HCl and HEPES buffers with addition of 1 M NaCl or 750 mM NaCl showed a significant increase in the stability of BLG. The stability changes of BLG due to salt ionic effect can be seen in Figure 4. There is little significance of stability effect dependent on the buffer system (HEPES vs Tris-HCl). Salt concentration is what is greatly altering the melt curves and causing stabilisation. The buffer screen tested had a highest salt concentration of 1 M in this instance, no higher concentrations were tested. This high salt concentration causes significant positive changes to stability for BLG.
Conclusion
Beta-Lactoglobulin is the most abundant protein found in bovine milk. It has undergone many stability and folding studies due to its importance in the dairy industry. Its physiological role is yet to be defined though it is hypothesised to be involved in the transport of molecules between parent and offspring [4]. The confirmation and stability of BLG has been linked greatly with altered pH conditions and buffer composition [5]. BLG notably alters oligomeric state at differing pHs, whether its monomeric or dimeric form affects stability would require further research approaches [6]. Our screen showed increased stability of BLG in not only citric acid but also low pH SPG, indicative of a preference for acidic buffers to maintain stability. The results shown corroborate with the published literature of BLG, whereby structure has been elucidated successfully in high salt acidic buffer screens, specifically, 3 M NaCl buffered at pH3.8 with sodium citrate [2]. The stability of proteins in an initial buffer condition screen has been directly linked with crystal formation success [1]. The DSF data reported shows success in being able to narrow down conditions before crystallisation screening trials. This confirmation of results outlines SUPR‑DSF as a suitable high throughput system to rapidly screen optimal buffer conditions for recombinant proteins. As many commercial buffer screens come in 96-well format, the SUPR-DSF 384-well format allows replicate screening within the same experimental set up. SUPR-DSF is a suitable system for buffer screening before downstream structural methods and as a primary screen before setting up of crystallography plates. The need to not add extrinsic dyes also aids in elucidation of true protein solubility results. Buffer screening for protein homogeneity should be considered an integral part of protein purification research.
Methodology
RUBIC Buffer Screen was dispensed into 384 well PCR microplate to a well volume of 18 µL, 4 replicates. Beta-Lactoglobulin was prepared at 5 mg⁄mL in water, 2 µL was dispensed in the previously prepared buffer screen plate to achieve a total well volume of 20 µL and protein concentration of 0.5 mg⁄mL. After dispensing, the plate was centrifuged for 30 seconds and sealed with an optically clear adhesive film.
The SUPR-DSF was set up to measure the fluorescence spectra from 10 °C to 105 °C with a 1 °C per minute ramp rate. Quantification of spectra and subsequent Tm was determined by the barycentric mean (BCM). The heat map was generated using mean of 4 replicates for each buffer condition. Methods and results for all samples are available on request.
References
[1] - J. Jancarik, R. Pufan, C. Hong, S.H. Kim, R. Kim. “Optimum solubility (OS) screening: an efficient method to optimize buffer conditions for homogeneity and crystallization of proteins”. Acta Crystallographica Section D: Biological Crystallography, Vol. 60(9), 1670-1673. 2004.
[2] - J. Loch, A. Polit, A. Górecki, P. Bonarek, K. Kurpiewska, M. Dziedzicka‐Wasylewska, K. Lewiński, “Two modes of fatty acid binding to bovine β‐lactoglobulin—crystallographic and spectroscopic studies”, Journal of Molecular Recognition, Vol. 24(2), 341-349. 2011.
[3] - S. Boivin, S. Kozak, R. Meijers, “Optimization of protein purification and characterization using Thermofluor screens”, Protein expression and purification, Vol. 91(2), 192-206. 2013.
[4] - S. Świątek, P. Komorek, G. Turner, B. Jachimska, “β-Lactoglobulin as a potential carrier for bioactive molecules”. Bioelectrochemistry, Vol. 126, 137-145. 2019.
[5] - L. Vijayalakshmi, R. Krishna, R. Sankaranarayanan, M. Vijayan, “An asymmetric dimer of β‐lactoglobulin in a low humidity crystal form Structural changes that accompany partial dehydration and protein action”. Proteins: Structure, Function, and Bioinformatics, Vol. 71(1), 241-249. 2008.
[6] - T. O. Yeates, A. Mc Pherson, “The structure of bovine β-lactoglobulin in crystals grown at pH 3.8 exhibiting novel threefold twinning”. Acta Crystallographica Section F: Structural Biology Communications, Vol. 75(10), 640-645. 2019.
