Formulation Screen of Trastuzumab Using the SUPR-DSF
Introduction
Successful formulation of biotherapeutic antibodies is fundamental to ensuring a stable and safe product that can withstand manufacture, storage, and transportation. This development process requires high throughput methods to screen a large combination of formulation conditions reliably and quickly to identify the best candidate and optimal conditions.
A widespread method used is thermal denaturation and derived parameters like Tm and Tonset to rank formulation conditions and excipients on their stabilising effects. Traditional label-free techniques for thermal denaturation have been limited in throughput by sample requirement concentrations and volumes, consumable costs, and measurement speed. This makes it challenging for formulation screening campaigns with hundreds or thousands of potential conditions.
In this report, we show the analysis of a thermal denaturation-based formulation screen of the commercially available therapeutic antibody Trastuzumab in 96 different conditions with the SUPR-DSF instrument. Along with screening the stabilising agents, confidence in the results is gained as there is consistency with both Differential Scanning Calorimetry and the formulation used for the commercial drug Herceptin®. This screen was directly measured in a single 384‑well microplate in less than 1.5 hours. This high‑throughput can be leveraged further through lab automation integration to screen thousands of samples per day.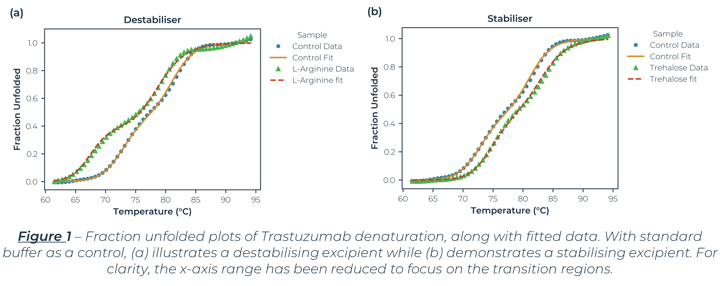
Results
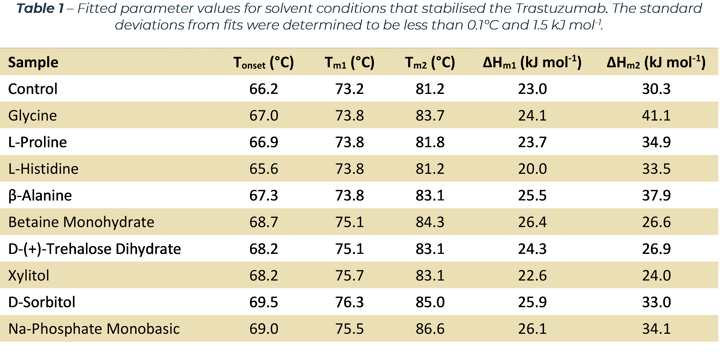
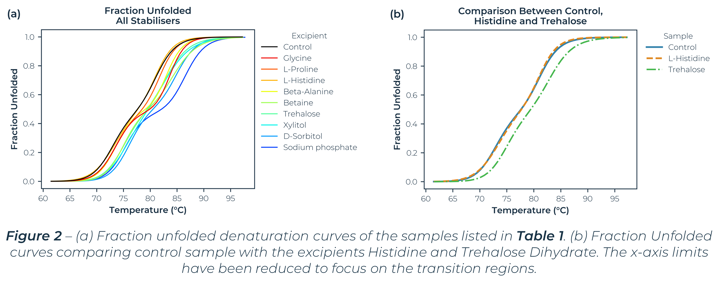
The denaturation curves measured (which Figure 1 and 2 are exemplars) exhibit two separate transition regions. Through Least-Squared fitting of the denaturation data, via a standard three‑state model[1], the Tm values, van 't Hoff enthalpy (ΔHm) and Tonset values were determined and are listed in Table 1. The table lists all the excipients that improved the stability of the antibody (compared to the control sample).
The three-state behaviour is consistent with DSC data for Trastuzumab[2] which reports two Tm values around 70°C and 82°C. Acknowledging sample and solvent variability, the Tm values are consistent with values reported elsewhere.[2] Along with similar Tm values, the SUPR-DSF data established that both histidine and trehalose dihydrate either did not hinder or improved the conformational stability of Trastuzumab, which are formulation constituents in the Trastuzumab containing drug Herceptin®.[3] These two points help demonstrate SUPR-DSF’s measurement consistency, all while offering superior throughput capabilities.
Conclusion
The SUPR-DSF from Protein Stable demonstrates high data quality, capable of quantifying multi‑domain unfolding events of the biotherapeutic antibody Trastuzumab under 96 formulation conditions in a single 1.5-hour measurement. The results are consistent with those reported by other lower throughput methods, and correctly identified excipients that are used in the marketed biotherapeutic formulation of Trastuzumab.
In addition, the intrinsic fluorescence based Differential Scanning Fluorimetry method of the SUPR-DSF can achieve these outcomes all while increasing throughput, convenience, and lowering consumables cost and sample consumption.
This already data rich and high throughput method can be leveraged further through integration with lab automation to screen well over a thousand conditions per day.
Methodology
Microplate Preparation
Trastuzumab was prepared at 1 mg⁄mL. The 96 different solvent conditions were obtained pre‑formulated in a deep well 96-well plate. Samples were prepared in the wells of a 384‑well, commercial polypropylene PCR microplate. The final antibody concentration of 0.1 mg⁄mL was achieved by dispensing 2 µL of antibody stock into 18 µL formulation solution to a final well volume of 20 µL. After dispensing, solution mixing was achieved by shaking the plate for 5 mins at 1500 rpm before being sealed with an optically clear adhesive film.
Thermal Denaturation Measurement
The SUPR-DSF was set-up to measure the fluorescence spectra of Trastuzumab samples from 20°C to 100°C, with a 1 °C per minute ramp rate. Quantification of the spectral shift, that is indicative of protein denaturation, was determined via the barycentric mean (BCM) calculation.
The thermal denaturation curves were fitted to a three-state model[1] via a Least-Squared regression algorithm, the fitted parameter values of which are listed in Table 1. Identification of better performing excipients was achieved by identifying samples with Tm1 and Tm2 values that were higher than the control sample. Methods and results for all samples are available on request.
References
[ 1] – J. Walters, S. L. Milam, A. C. Clark. “Practical Approaches to Protein Folding and Assembly: Spectroscopic Strategies in Thermodynamics and Kinetics”. Methods in Enzymology. Vol. 445, p. 1‑39. 2009.
[2] – K. M. Hutterer, A. Polozova, S. Kuhns, H. J. McBride, X. Cao, & J. Liu. “Assessing Analytical and Functional Similarity of Proposed Amgen Biosimilar ABP 980 to Trastuzumab”. BioDrugs, vol. 33(3), p. 321–333. 2019.
[3] – Genentech Inc. “Herceptin (Tratuzumab) Package Insert”, U.S. Food and Drug Administration, 1998.
